About Us

The Dry Bean Breeding & Computational Biology Program at the University of Guelph is a cornerstone of agricultural innovation. Since 1977, we have been dedicated to advancing dry bean varieties. We combine decades of breeding expertise with modern computational tools to deliver resilient, high-quality beans for Ontario growers and global markets.
Our History
The Field Bean Breeding Program began in 1977, initially focusing on white bean development with support from the Ontario Ministry for Agriculture, Food, and Rural Affairs, the Ontario White Bean Producer’s Marketing Board, and the Ontario Coloured Bean Growers (merged into the Ontario Bean Growers in 2013). Our first release, Ex Rico 23 (1980), a navy bean, marked the start of our legacy. A major milestone followed in 2001 with OAC Rex, Canada’s first Common Bacterial Blight (CBB)-resistant navy bean, bred from an interspecific cross with a wild species. Both varieties were honored in the 2012 Seed of the Year Award.
In 2009, we partnered with Agriculture and Agri-Food Canada (AAFC), the Ontario Bean Producers Marketing Board, and the Ontario Coloured Bean Growers to form a consolidated breeding program at Guelph. This collaboration ended in 2014, leaving Guelph as Ontario’s sole bean breeding hub until AAFC Harrow resumed efforts in 2016. Today, we work alongside AAFC and Guelph researchers to conduct the Ontario Registration and Performance Trials, ensuring new varieties thrive in local conditions.
“From pioneering varieties to shaping sustainable agriculture, our history reflects a commitment to excellence.”
Our Mission Today
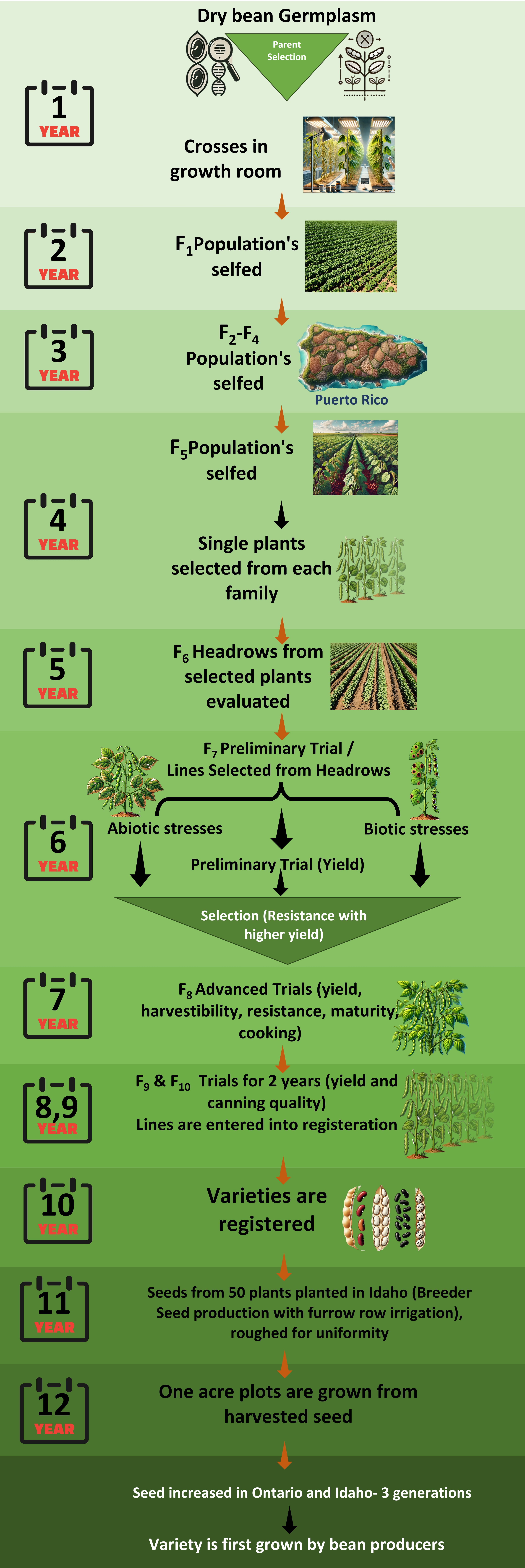
Our breeding program implements a modified single-plant selection system designed to efficiently identify elite genotypes from thousands of segregating individuals across multiple bean market classes. Each year, we:
– Make 200–250 crosses spanning navy, black, cranberry, kidney (light, dark, and white), pinto, yellow, and adzuki beans to introduce new combinations of alleles.
– Advance populations from F2 to F5, maintaining broad genetic diversity until the onset of intensive selection.
– Initiate single-plant selection at the F5 stage, using a two-step process:
Round 1 (Field): Select up to 5,000 single plants based on plant architecture, maturity, lodging resistance, disease reaction, pod load, and overall visual appearance.
Round 2 (Lab): From these, select up to 3,500 single plants based on seed traits including colour, size, shape, brightness, uniformity, and market-class characteristics.
– Grow and evaluate headrows the following year, discarding off-types and retaining only uniform, true-breeding lines.
– Progress superior lines into preliminary and advanced yield trials over multiple locations, assessing >50 agronomic, disease, and quality traits per line.
– Conduct two years of official Ontario Registration and Performance Trials, ensuring agronomic merit, stability, and market acceptance.
– Collaborate closely with OPCC and other provincial partners to advance, release, and register new cultivars for Ontario growers.
This multi-stage pipeline enables continuous improvement and rapid development of high-performing cultivars tailored to diverse Ontario environments and end-use requirements.
Breeding Pipeline
Figure 1: Workflow and co-ordination of information in the bean breeding and computational biology program at the University of Guelph.
Objectives for Ontario Growers
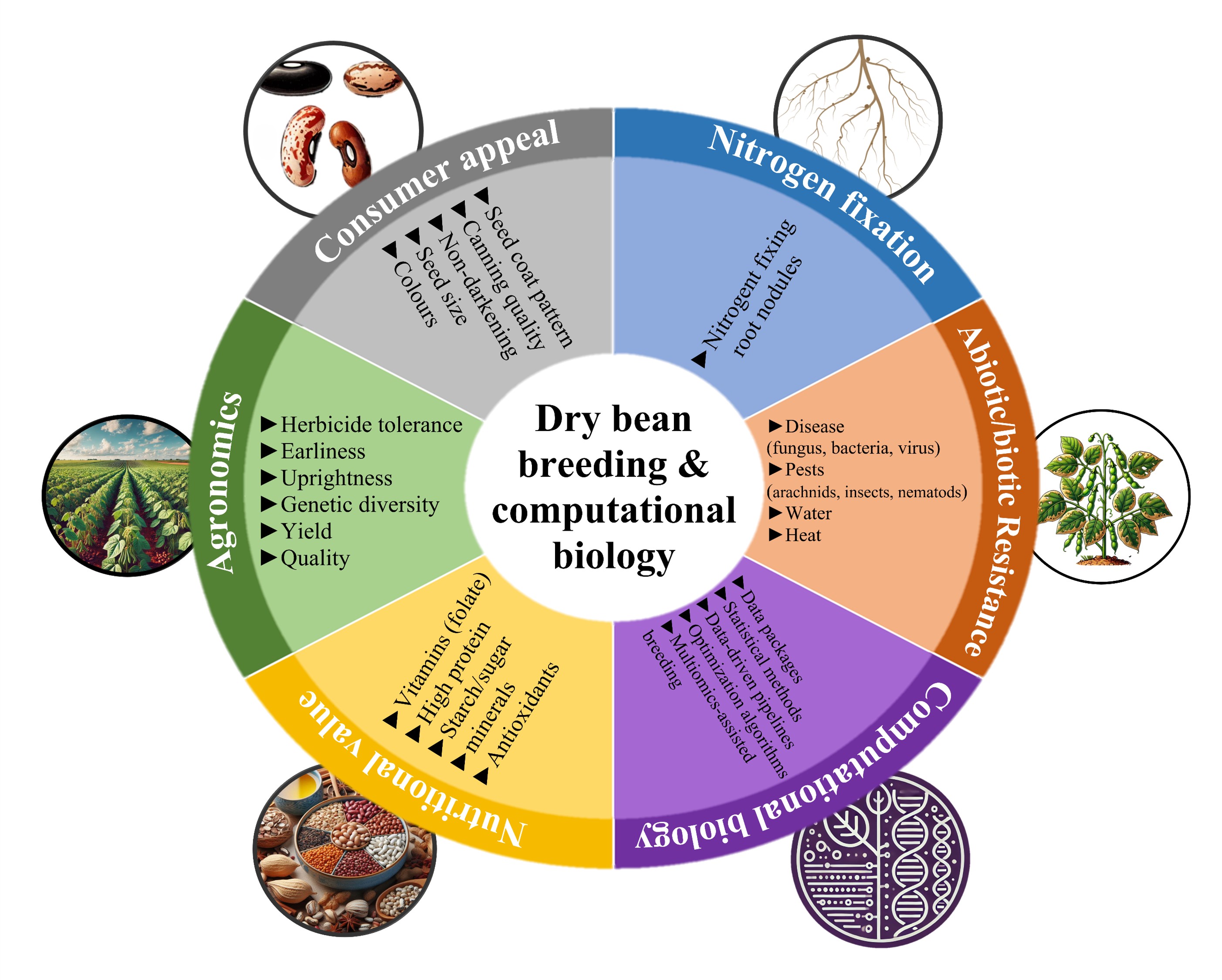
Our work is guided by four key goals:
- Yield Improvement: Maximizing productivity to boost farmer profitability.
- Stress Resistance: Enhancing resilience against pests, diseases, nematodes, drought, and heat.
- Nutritional & Quality Enhancement: Improving nutritional content and cooking quality for healthier diets.
- Market Class Diversification: Developing diverse bean types to meet consumer and trade demands.
Program Goals
Figure 2: Bean breeding objectives at the University of Guelph.
Our Approach
We blend conventional breeding with modern innovations:
- Genomic Selection: Leveraging genetic markers for precise trait selection.
- Phenomics: Using high-throughput imaging to evaluate plant performance.
- Computational Biology: Applying advanced analytics to optimize breeding decisions.
Our collaboration with AAFC and Guelph researchers ensures varieties are tailored to Ontario’s climate and soil, tested rigorously through the Ontario Registration and Performance Trials.
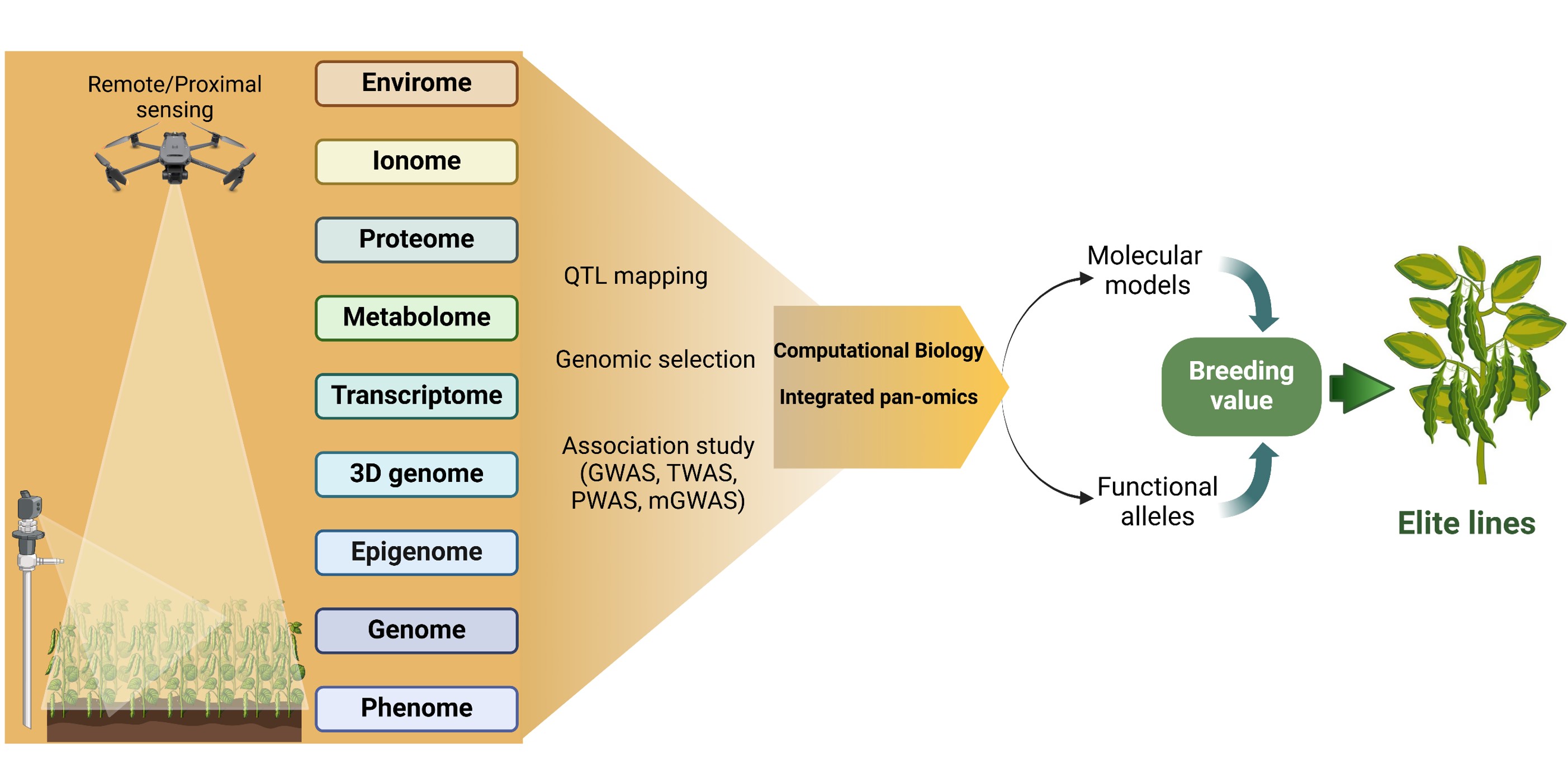
Figure 3: Multi-omics (genomics, 3D genomics, transcriptomics, epigenomics, proteomics, metabolomics, ionomics, enviromics, phenomics)-assisted dry bean breeding. Advances in next-generation sequencing, high-throughput phenotyping, and computational biology will enable a more comprehensive understanding of breeding value and facilitate the introduction of elite lines in dry beans.The figure was created using BioRender.com.
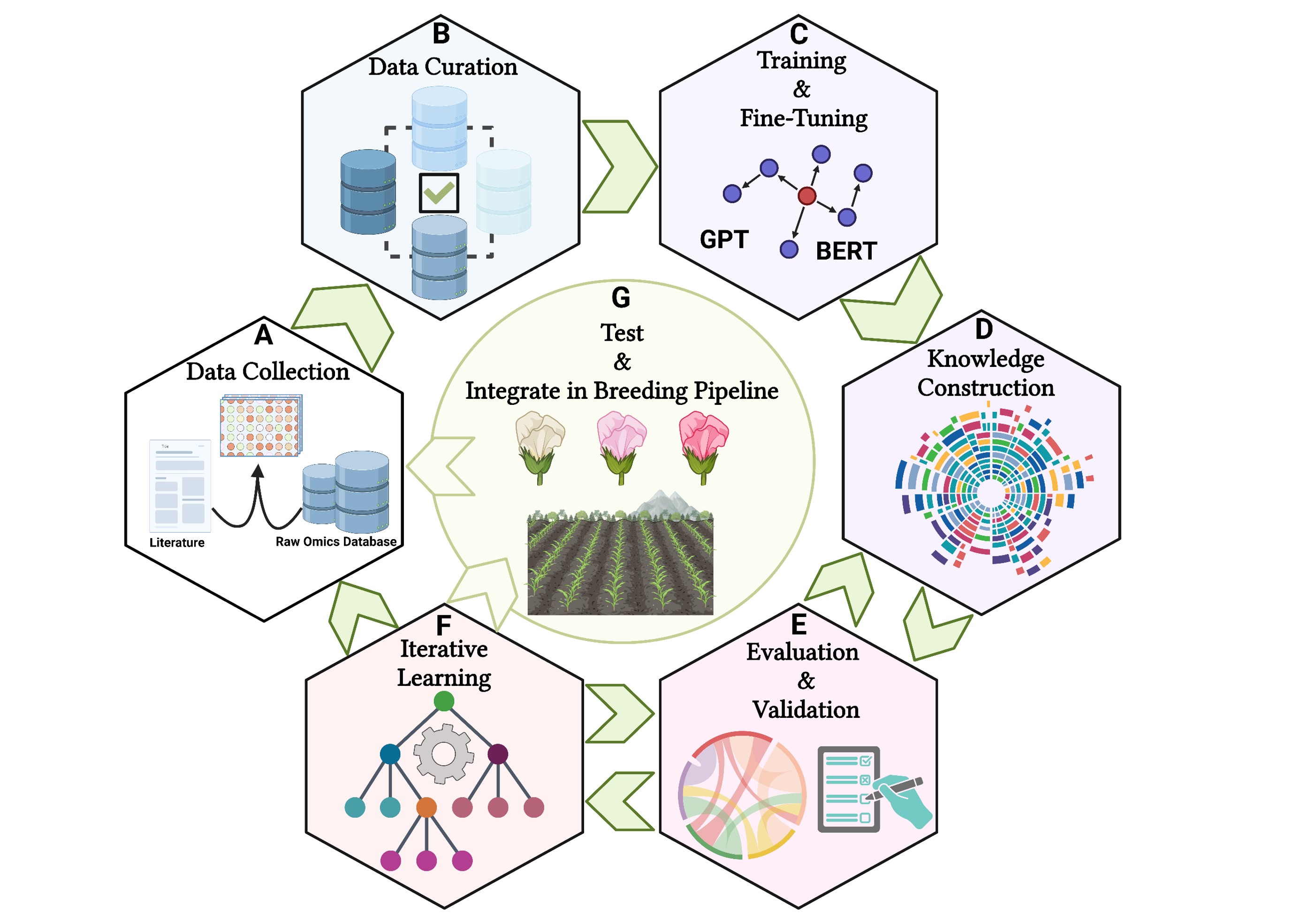
Figure 4: A schematic picture of utilizing LLMs in plant breeding area. A) Collecting diverse datasets from scientific literature, multi-omics repositories, and breeding records, B) Standardizing text and annotating multi-omics data during preprocessing, C) Choosing a pre-trained LLM, fine-tuning it with plant breeding-specific texts, and using multi-modal methods to integrate text and structured data, D) Leveraging the LLM to build knowledge graphs that illustrate the relationships between multi-omics, traits, and environmental factors, E) Establishing performance metrics and refining outputs with input from breeders and biologists, F) Creating feedback loops to continuously assess results, and G) Assisting plant breeders in developing data-driven strategies by prioritizing multi-omics and traits for field trials based on their yield, quality, and adaptability. The figure was created using BioRender.com.